Key Enzymes at the DNA Replication Fork
DNA replication is an important biological process that ensures accurate transmission of genetic information from one generation to the next. Central to this process is the replication fork (ScienceDirecte), a Y-shaped structure that forms when the DNA double helix is unwound. This fork acts as a site for the coordinated action of several key enzymes, each playing a vital role in copying the genetic material. Understanding these enzymes is essential for comprehending how cells maintain genetic stability and division.
1. DNA Helicase
The first step in DNA replication involves unwinding the double helix to expose the individual strands. This is achieved by DNA helicase, a motor protein that moves along the DNA, breaking the hydrogen bonds between complementary base pairs. As helicase unwinds the DNA, it creates two single-stranded templates for replication. The activity of helicase is essential for maintaining the replication fork's progression and preventing DNA tangling.

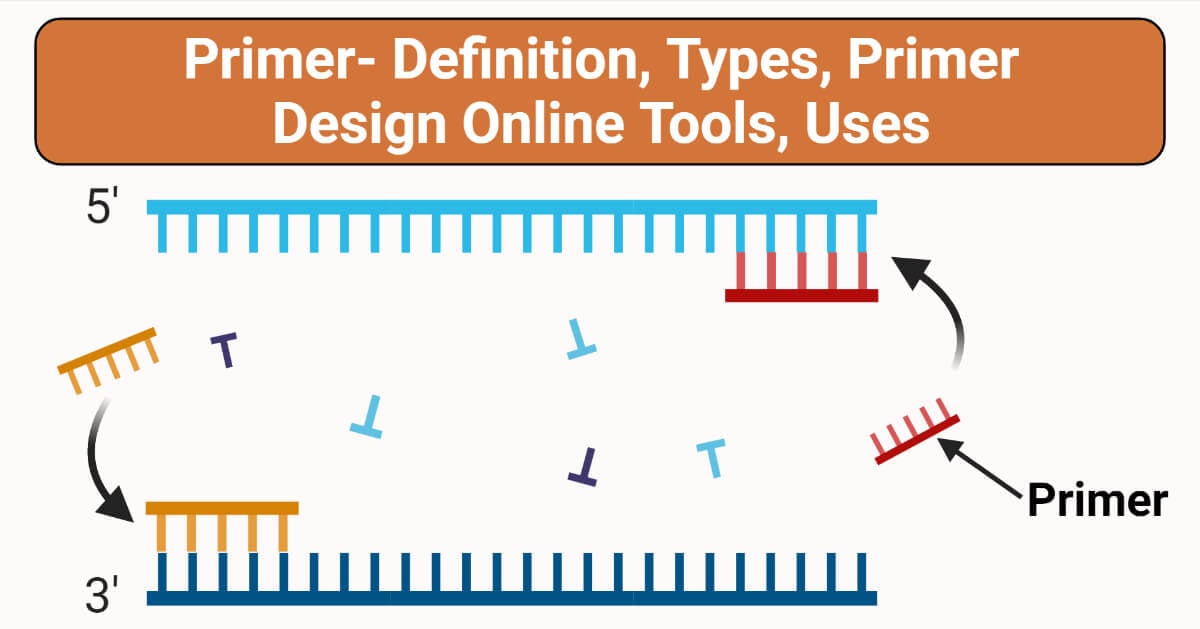
2. DNA Primase
Once the DNA strands are separated, the next enzyme to join the process is DNA primase. This enzyme synthesizes short RNA primers that serve as starting points for DNA polymerases. Since DNA polymerase can only add nucleotides to an existing strand, primase ensures that a primer is in place to kickstart replication. The primers are later removed and replaced with DNA.
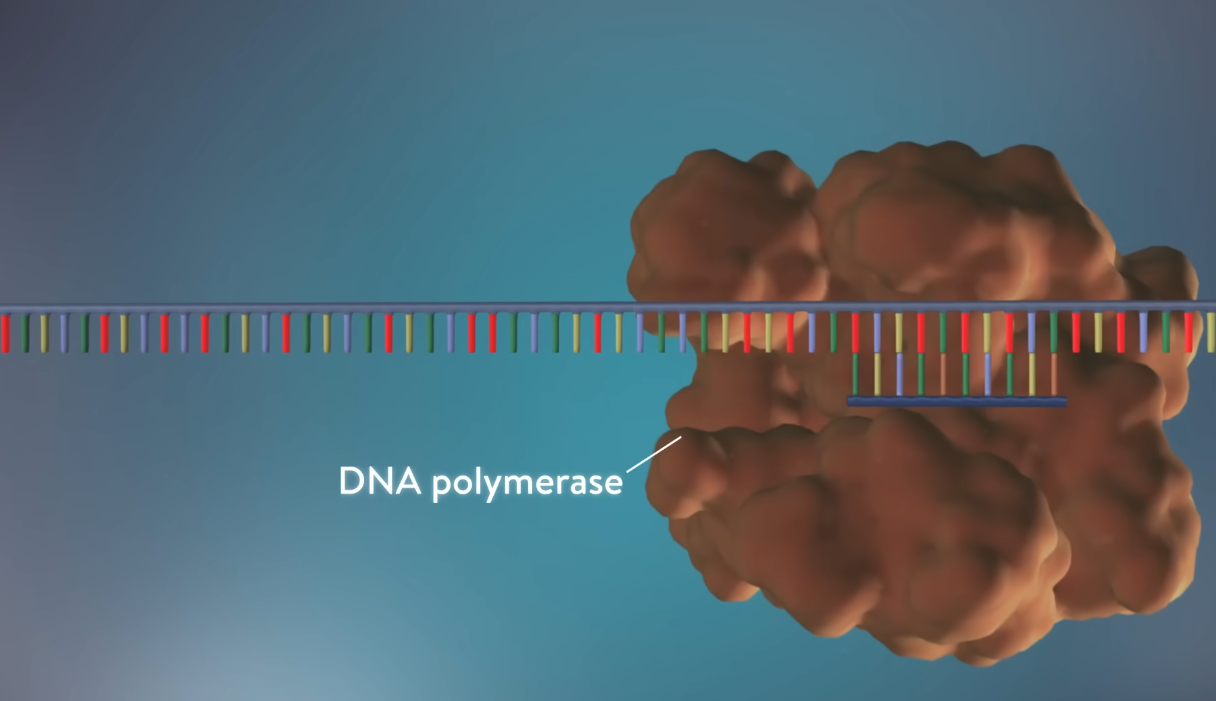
3. DNA Polymerase
At the heart of DNA replication is DNA polymerase, the enzyme responsible for synthesizing the new DNA strand. DNA polymerase works in a 5' to 3' direction, adding nucleotides to the growing strand by matching them with their complementary bases on the template strand. The enzyme also has a proofreading function, ensuring that any errors in nucleotide pairing are corrected, maintaining the accuracy of replication.
4. Single-Stranded Binding Proteins (SSBs)
During DNA replication, the double helix is unwound to expose the individual strands, creating single-stranded DNA regions. These exposed strands are highly susceptible to re-annealing, degradation, or formation of secondary structures, which can interfere with the replication process. SSBs stabilize the DNA by preventing the strands from folding back onto themselves or pairing with complementary sequences. This binding ensures that the single-stranded regions remain available for the replication machinery, particularly for the action of DNA polymerases. Moreover, SSBs protect the single-stranded DNA from degradation by nucleases, which could otherwise cleave the DNA, ensuring the preservation of the genetic material during replication. By performing these critical functions, SSBs maintain the integrity and efficiency of the replication fork, allowing for accurate DNA synthesis.
5. DNA Ligase
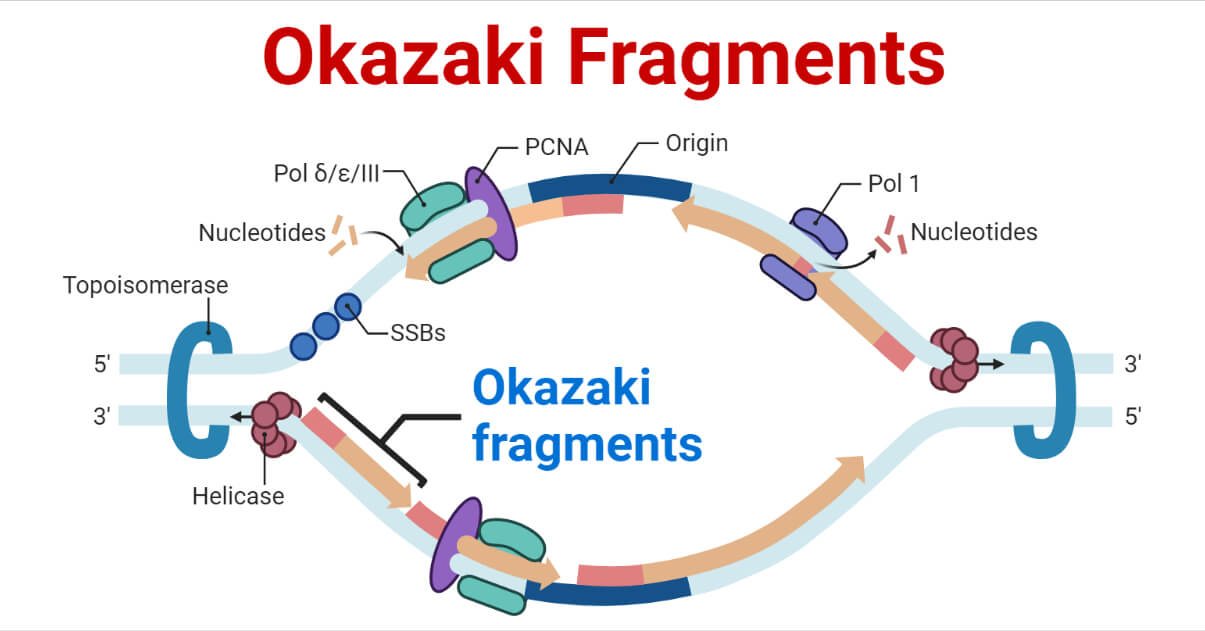
After DNA polymerase synthesizes the new strands, it leaves behind small gaps, especially on the lagging strand where replication occurs in short bursts known as Okazaki fragments. DNA ligase plays a important role in sealing these gaps by forming phosphodiester bonds between adjacent nucleotides. This enzyme ensures that the newly synthesized DNA is continuous and intact, completing the replication process.
Okazaki fragments