Description
MagSi-DNA | MD02017
I. Intended use
MagSi-DNA beads are ideal for purification or isolation of nucleic acids from various sources. The magnetic nanoparticles are intended as a solid phase extraction tool for custom buffer systems based on chaotropic as well non-chaotropic binding principles, and can be used for developing your own nucleic acid isolation and extraction methods, such as:
● Isolation of genomic, mitochondrial, or viral DNA from whole blood, cell lysates, human, animal, or plant tissue; isolation of RNA
● Isolation of genomic, plasmid, or phage DNA from bacterial cultures and bacteria from clinical samples (blood, stool, swabs, etc.)
● Clean-up of DNA from enzymatic reactions (restriction digestions, ligations) or chromatin immunoprecipitation (ChIP) procedures to remove excess primers, nucleotides, enzymes, salts, buffers and other substances that are unwanted in downstream applications MagSi-DNA beads are magnetic silica beads with a highly dense magnetic core of iron oxide. Due to their magnetic properties, the beads typically collect within 10 seconds in a magnetic field. Their small particle size offers a very large active surface area and high binding capacity for nucleic acids. These particles are applicable in both manual and automated processes, but continuous shaking is needed because of their fast sedimentation.
II. Principle
MagSi-DNA reversibly binds DNA and other nucleic acids under sample- and buffer-specific conditions. A solution containing DNA (e.g.lysate) is combined with the beads and an application-specific binding buffer. After incubation, nucleic acids are bound to the silica surface. By applying a suitable magnet to the container (tube/deepwell
microplate) the bead pellet is separated from the sample mixture. Unwanted components are further removed by washing steps in a selection of buffers (alcohol/water solutions). Finally, nucleic acids are released in DNase/RNase-free water or buffer solution (e.g. Tris, TrisEDTA, pH~8).
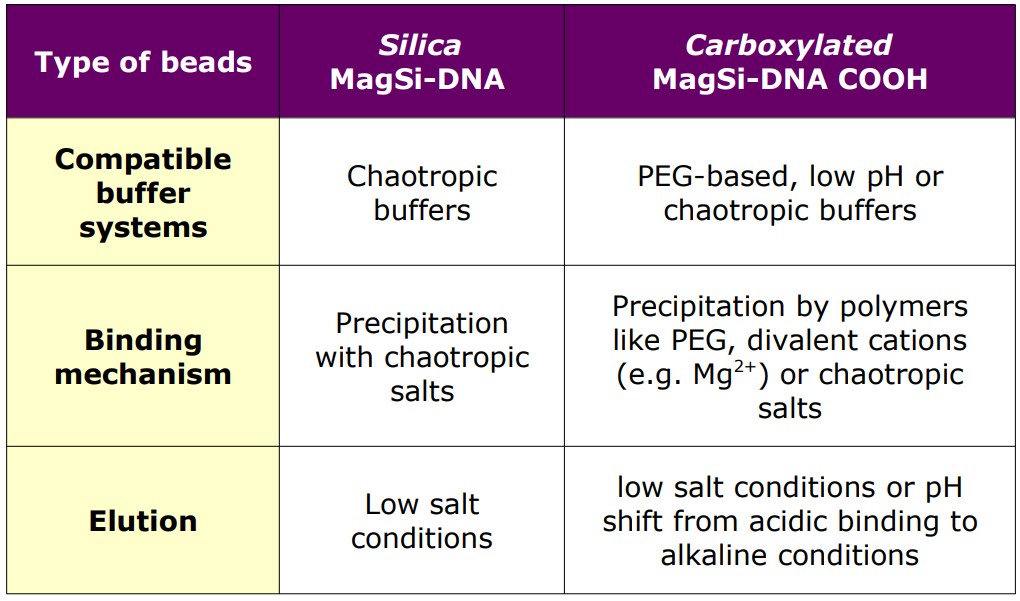
Silica and carboxylated (COOH) surfaces, but also nucleic acids, arenegatively charged at neutral or basic pH, while both are also hydrated. For a chaotropic binding mechanism of DNA to particles,dehydration is needed. This can be achieved by for instance alcohol, and by agents such as guanidinium salts. Negative charges on the bead surface and the nucleic acid backbones are bridged by divalent cations. This can be reversed by a water solution.
For washing, mostly alcohol/water mixtures are used, which will keep the DNA in dehydrated form and bound to the beads. To reduce premature elution of DNA, salts can be added to the washing solution. Elution takes place in a low-salt conditions. Non-chaotropic systems may use binding mechanisms with specific
pH conditions, or binding by polyethylene glycol precipitation.
Silica & Carboxylated MagSi-DNA beads
Optimal binding conditions differ for beads with silica or with carboxylated surfaces. In Table 1 below, some of the practical differences between the 2 types of beads are shown. (To develop a new application it is recommended to try both types in parallel!
Table 1: Differences between silica (MagSi-DNA) and carboxylated (MagSi-DNA COOH) beads